Page 661 - TNFlipTest
P. 661
Toronto Notes 2019 Introduction to Genetics Pedigrees
• diagramsthatshowthepattern/distributionofphenotypesforageneticdisorderwithinafamily,often across multiple generations
Medical Genetics MG3
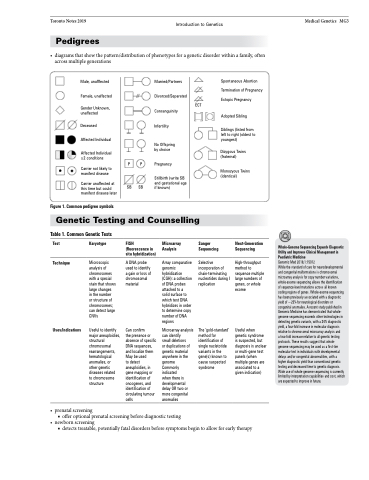
Male, unafffected
Female, unaffected
Gender Unknown, unaffected
Deceased
Affected Individual
Affected Individual ≥2 conditions
Carrier not likely to manifest disease
Carrier unaffected at this time but could manifest disease later
Figure 1. Common pedigree symbols
Married/Partners Divorced/Separated Consanguinity Infertility
No Offspring by choice
P Pregnancy
ECT
Spontaneous Abortion Termination of Pregnancy Ectopic Pregnancy
Adopted Sibling
Siblings (listed from left to right (oldest to youngest)
Dizygous Twins (fraternal)
Monozyous Twins (identical)
P
SB SB
Stillbirth (write SB and gestational age if known)
Genetic Testing and Counselling
Table 1. Common Genetic Tests
Test
Technique
Uses/Indications
Karyotype
Microscopic analysis of chromosomes with a special stain that shows large changes
in the number or structure of chromosomes; can detect large CNVs
Useful to identify major aneuploidies, structural chromosomal rearrangements, hematological anomalies, or
other genetic diseases related to chromosome structure
FISH (fluorescence in situ hybridization)
A DNA probe used to identify a gain or loss of chromosomal material
Can confirm
the presence or absence of specific DNA sequences, and localize them May be used
to detect aneuploidies, in gene mapping or identification of oncogenes, and identification of circulating tumour cells
Microarray Analysis
Array comparative genomic hybridization (CGH): a collection of DNA probes attached to a solid surface to which test DNA hybridizes in order to determine copy number of DNA regions
Microarray analysis can identify
small deletions
or duplications of genetic material anywhere in the genome
Commonly indicated
when there is developmental delay OR two or more congenital anomalies
Sanger Sequencing
Selective incorporation of chain-terminating nucleotides during I replication
The ‘gold-standard’ method for identification of single nucletotide variants in the gene(s) known to cause suspected syndrome
Next-Generation Sequencing
High-throughput method to sequence multiple large numbers of genes, or whole exome
Useful when genetic syndrome is suspected, but diagnosis is unclear or multi-gene test panels (when multiple genes are associated to a given indication)
Whole-Genome Sequencing Expands Diagnostic Utility and Improves Clinical Management in Paediatric Medicine
Genomic Med 2016;1:15012
While the standard of care for neurodevelopmental and congenital malformations is chromosomal microarray analysis for copy number variations, whole-exome sequencing allows the identification of sequence-level mutations across all known coding regions of genes. Whole-exome sequencing has been previously associated with a diagnostic yield of ~25% for neurological disorders or congenital anomalies. A recent study published in Genomic Medicine has demonstrated that whole- genome sequencing exceeds other technologies in detecting genetic variants, with a 34% diagnostic yield, a four-fold increase in molecular diagnosis relative to chromosomal microarray analysis and
a two-fold increase relative to all genetic testing protocols. These results suggest that whole- genome sequencing may be used as a first-tier molecular test in individuals with developmental delays and/or congenital abnormalities, with a higher diagnostic yield than conventional genetic testing and decreased time to genetic diagnosis. Wide use of whole-genome sequencing is currently limited by interpretation capabilities and cost, which are expected to improve in future.
• prenatalscreening
■ offer optional prenatal screening before diagnostic testing
• newbornscreening
■ detects treatable, potentially fatal disorders before symptoms begin to allow for early therapy